One of the things that makes the initial learning curve of R so steep is that R doesn’t have a point and click, menu based, graphical user interface. Learning R is to some degree synonymous with learning the R programming language.
Don’t let the words ‘programming language’ stop you here. While it might seem daunting at first – especially if this is your first contact with a programming language – it’s honestly not that bad.
We’ve already explored some of the way the R command line works in the last two posts. You’ve seen that R can be used like a calculator to reduce mathematical expressions.
In many ways, the rules of even basic mathematics can be viewed as a programming language by which you communicate commands to a program like R. The expression of ‘3+4’ takes on a very specific form in order to convey the intention of adding the number 3 to the number 4. Entering the form ‘+34′ or ’34+’ gives incorrect responses if the goal is to add 3 to 4.
While the expression ‘4+3’ is equivalent here, the same isn’t true for all the basic operators (‘3/4’ is not equivalent to ‘4/3’). The way we form these expressions adheres to a ruleset that you probably haven’t had to think of since grade school. The reason is exposure and practice. Use a programming language long enough and it will start to feel as natural as addition and subtraction.
In addition to calculations, we’ve already seen a few other basic commands in the first few posts. Among these, and one of the commands you will use the most in R, is ‘<-‘
Recall that the command:
x<-3
Stores a value of 3 into the variable x. The reason that you’ll use this frequently is that it is exceptionally powerful. Being able to store values to variables is foundational to many of the more complex things you’ll do in R.
Now, it’s worth noting that this is not the only way to store values to a variable. The following are both equivalent to the above:
3->x
x=3
While using the forward arrow (‘->’) might not be particularly enticing, it’s easy to get into the mindset that ‘=’ would be an easier way to handle variable assignment. While there’s nothing stopping you, the general convention is to use ‘<-‘.
This isn’t entirely arbitrary. While I can’t say for sure why this convention was established, there are a number of reasons why it makes sense. The most convincing of these is that the equal sign will be used for a number of other things, and there’s simply no reason to use it here. By saving it for tests of equivalence (‘==’) and definition of parameters within functions we can make things easier down the line.
Since we’re talking about it, you can use ‘==’ as a test of equivalence in R, and R will return a value of TRUE or FALSE depending on if the two elements being equated are equal or not. Try it with:
3==3
3==4
1+1==2
1+1==4
This will be particularly useful later, but is really just as simple as that at the moment. Store that one away, and just use ‘<-‘ instead of ‘=’ to assign values to variables.
Moving on, we’ve seen that we can use the function c() to group a number of values as one object. The most frequent use of c() is with variable assignment above, to create one variable that contains a number of elements. This isn’t always necessary, though, depending on what you’re looking to do.
If you simply type c(1,2,3,4) into the command line, R will, not surprisingly, return the values:
1 2 3 4
We could store those values into a variable with the command:
x<-c(1,2,3,4)
But, even without variable assignment, we could use c() as part of a mathematical expression. Suppose you wanted to convert a group of temperatures from Celsius to Fahrenheit. The formula for conversion is:
C*1.8+32=F
If we just type the first part of this expression into R, it will give us the temperature value in Fahrenheit. You can try this by typing the expression:
10*1.8+32
R should return a value of 50 (degrees).
If we wanted to check a number of temperatures all at once, we could take advantage of c() instead of running the same equation multiple times.
If you type the expression:
c(0,5,10,15,20,25)*1.8+32
R should return the following values:
32 41 50 59 68 77
Again, it might make more sense to store your Celsius values into a variable, which is how you’d normally see this done. That is:
x<-c(0,5,10,15,20,25)
x*1.8+32
This should give the same result, and if you wanted you could even store that set of results as a variable by altering the second line to:
y<-x*1.8+32
Now, say we wanted to do the same thing, but instead of going from 0 to 25 we wanted to go from 0 to 100, in steps of 5. We could sit down and write out all the values, but there’s an easier way by using the function seq().
I’ve called c() a function above, without really going into what that means. A function is basically a command that takes some number of inputs and returns a result. The function c() takes all the values inside it and combines them into one set of values. The function seq() is only slightly more complicated.
If you simply type:
seq(10)
R will output the values:
1 2 3 4 5 6 7 8 9 10
If we put in 100, R will give us the values from 1 to 100. There is a lot we can start to learn about R from this.
One of the main things we can learn is that if we forget what a function in R does, we can use the function help() to find out. In this case, by typing:
help(“seq”)
This pulls up a help file on this function, and defines the parameters that the function accepts. There a number of parameters, and they all have defaults. We can interpret this from the following in the help file:
seq(from = 1, to = 1, by = ((to – from)/(length.out – 1)), length.out = NULL, along.with = NULL, …)
Forget about the later parts for now, but take note that the general form is:
seq(from,to,by)
If we only give one value, R interprets it to be the ‘to’ value, as that’s really all that can be done with one value (on the assumption that from and by are both 1 by default). We can add other values, and if we want to be organized we can even explicitly define them:
seq(from=1,to=10)
This should give the same output as seq(10) above, and will also give the same output as seq(1,10). It’s a good habit to explicitly define parameters while you’re first learning R, and a great habit to do it later. Fortunately or unfortunately, it’s not mandatory.
Keep in mind that what we’re looking to do is produce a set of numbers from 0 to 100 by steps of 5. That language is pretty close to the language that seq() can understand, so all we have to do is tweak it into the correct form:
seq(from=0,to=100,by=5)
This should give you the output:
0 5 10 15 20 25 30 35 40 45 50 55 60 65 70 75 80 85 90 95 100
Again, you can explicitly set those parameters by identifying each parameter that you’re setting, or just take advantage of the fact that the seq() function expects them in a certain order. That is, the following line is equivalent to the above:
seq(0,100,5)
We can now incorporate this into our earlier expression without having to use a c() function:
seq(0,100,5)*1.8+32
If you’ve been paying attention to the temperatures, the endpoints of the output shouldn’t be too shocking:
32 41 50 59 68 77 86 95 104 113 122 131 140 149 158 167 176 185 194 203 212
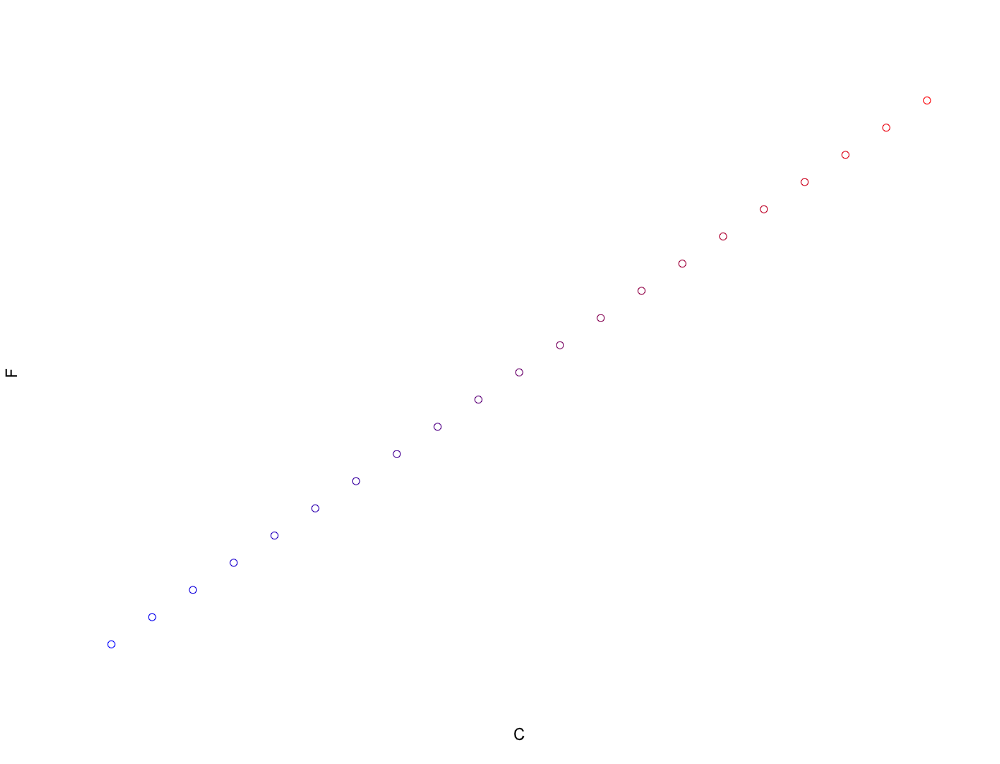
You might notice that each of these numbers is separated by 9 degrees Fahrenheit. That’s no coincidence – I’ve been using the conversion of:
C*1.8+32
1.8 can also be expressed as 9/5, so another way to write it is:
C*(9/5)+32
That is, every time the temperature goes up 5 degrees on the Celsius scale, it equates to a 9 degree temperature increase on the Fahrenheit scale. That’s what we’re seeing in our data above. Once we’re in Fahrenheit scaled units we have to add 32 degrees to account for the shift between the zero anchor point of the scales. It’s as simple as that.
Obviously, seq() can be used for a lot of different things. Play around with it, and see what you can create. Or, try to figure out the code below:
C<-seq(0,100,5)
F<-C*1.8+32
colFunction<-colorRampPalette(c(“blue”,”red”))
plot(C,F,col=colFunction(21))



 I am here to reassure you. You may be intimidated by this, and that is perfectly fine. This may very well be the first time you have encountered a command line interface. That is okay. You may be searching for other things you need to do right now instead of learning R, or deciding whether you want to just maybe worry about this tomorrow. Stick with me; today is the day.
I am here to reassure you. You may be intimidated by this, and that is perfectly fine. This may very well be the first time you have encountered a command line interface. That is okay. You may be searching for other things you need to do right now instead of learning R, or deciding whether you want to just maybe worry about this tomorrow. Stick with me; today is the day.

 You can see that each of my calculations were executed in sequence, giving the answers of 7, -1, 0.75, and 12 to the statements of 3+4, 3-4, 3/4, and 3*4.
You can see that each of my calculations were executed in sequence, giving the answers of 7, -1, 0.75, and 12 to the statements of 3+4, 3-4, 3/4, and 3*4.